Research in the Anslyn Group
Broadly speaking, our group focuses on physical organic, supramolecular, and materials chemistry. Using mechanistic insights and knowledge of photophysics, we devise sensing systems for real-life applications. While many of the sensors involve supramolecular interactions with analytes, we also exploit reversible covalent bonding in many cases. In this regard, we have generated a suite of reactions that can all occur simultaneously in the same solution with no crossover, which we refer to as Tunable Orthogonal Reversible Covalent (TORC) bonds. These TORC bonds are exploited reactions for material applications, polymer synthesis, complex assembly formation, and self-replicating oligomers. In addition, we have started a field that is a melding of computer science with organic chemistry, where information is stored in sequence-defined oligourethanes, whose sequences are read using a chain-end degradation routine, and subsequently, computer programs that recapitulate the data. We are moving these studies toward reading the sequences directly on integrated circuits, and manipulation of the information with exogenous chemistry. Finally, we have active collaborations with the Ellington group for making sustainable materials as well as mimicking life processes, and the Marcotte group for generating single molecule peptide sequencing routines.
Sequence Defined Oligourethanes
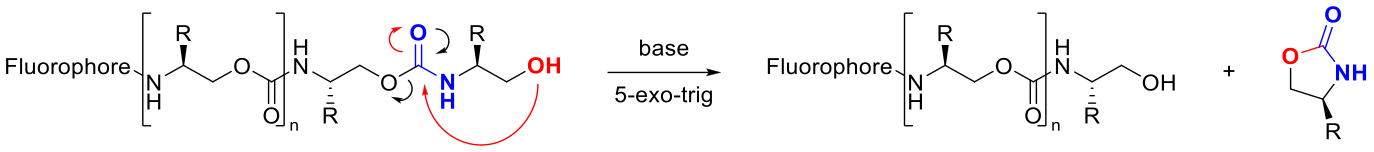
Sequence-defined polymers, such as DNA, have gained attention for their potential in biomimetics, catalysis, and information storage. Our group has developed the solid phase synthesis of sequence-defined oligourethanes (sd-OU) using commercially available amino alcohols. We have shown that sd-OU self-sequence in a controlled manner through 5-exo-trig cyclization of the urethane backbone, which can be monitored with LC-MS. We then applied this self-sequencing ability to information storage. As a proof of concept, a passage from Jane Austen’s Mansfield Park was encoded into hexadecimal through the sequence of –R functional groups in sd-OUs and then decoded by a third party with full accuracy. To further expand upon this work in information storage, we are synthesizing longer sequence-defined polyurethanes, thereby increasing the storage capacity of each molecule, as well as investigating techniques to increase the speed of information retrieval. To demonstrate the versatility of sd-OU for information storage in languages other than English, we are encoding information in pictographic languages, such as Mandarin. Lastly, we are using sd-OU to study enzymatic degradation of polyurethane plastics.
Related Publications:
ACS Cent. Sci. 2022, 8, 8, 1125-1133.
High-throughput enantiomeric excess determination
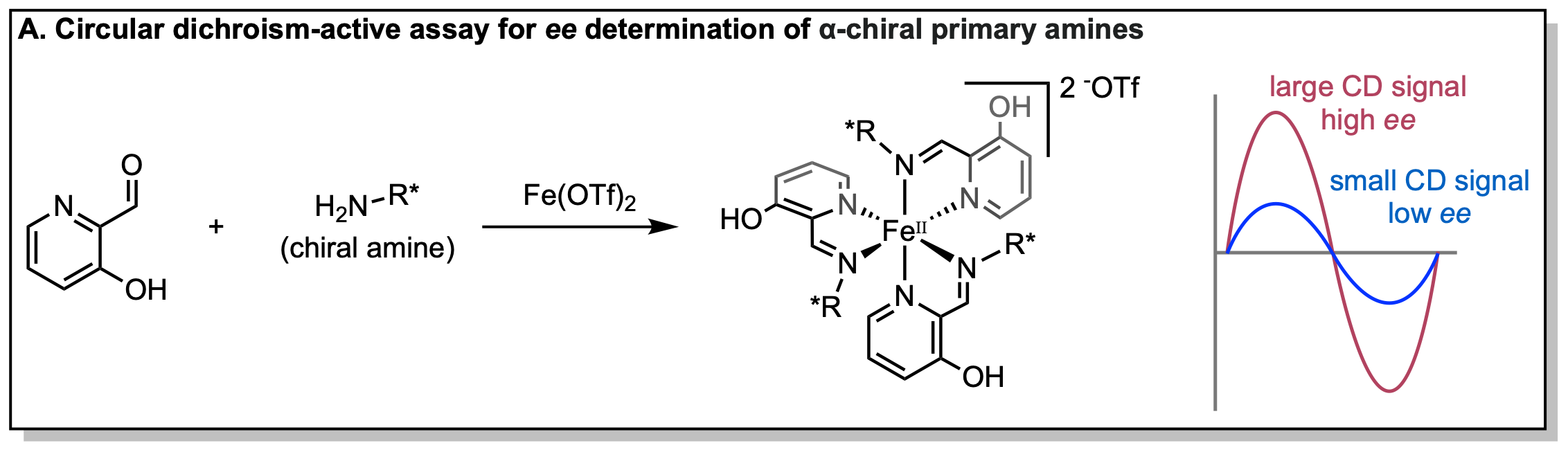
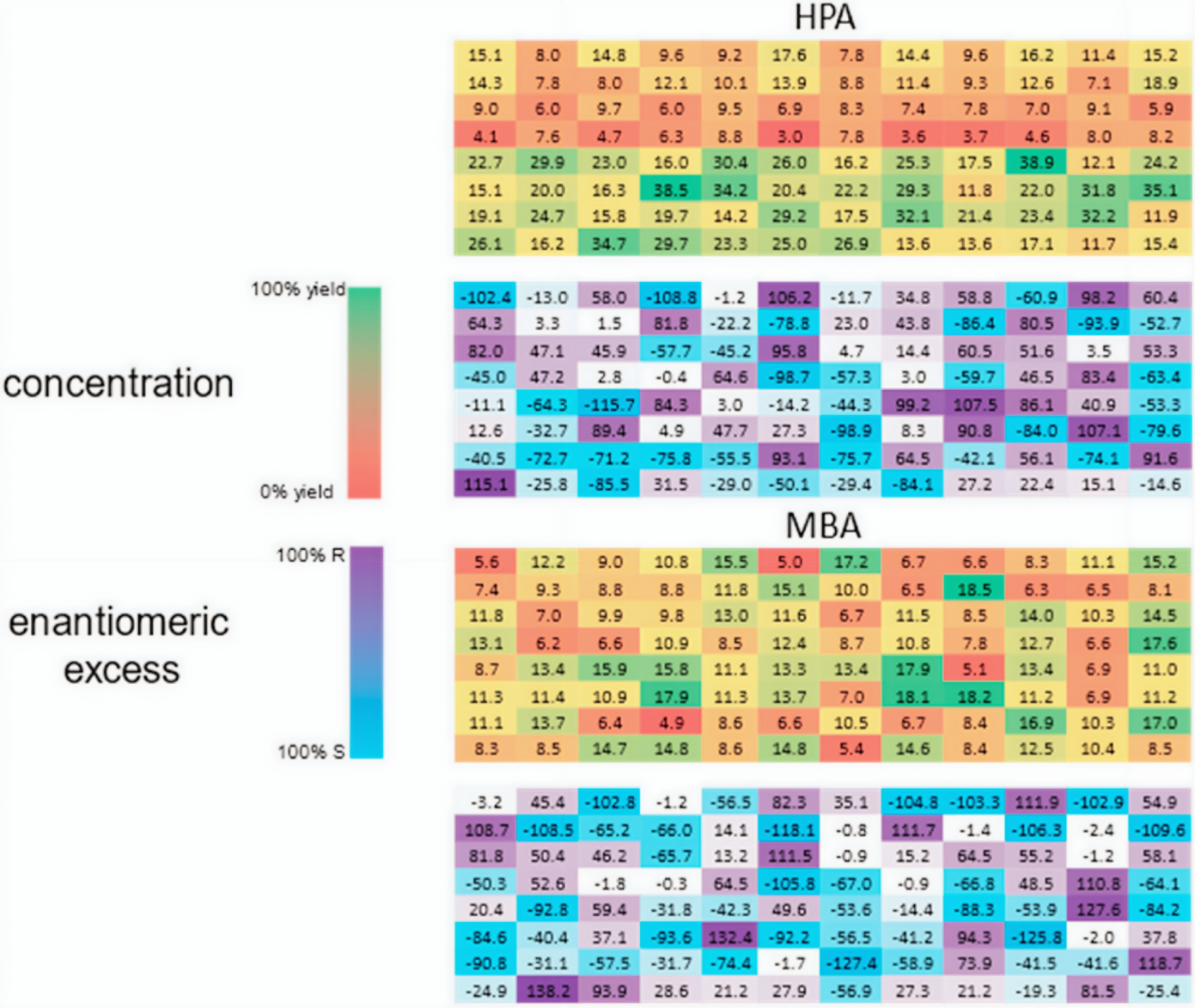
High-throughput experimentation (HTE) is a major focus of our group. HTE technologies in chemistry settings are often used for asymmetric reaction discovery and optimization in parallel. Although the methods for conducting reactions on such a scale is mature, the analysis of yield and enantiomeric excess (ee) is usually determined by chiral high-performance liquid chromatography (HPLC) with which reactions are analyzed sequentially. Our group and others have addressed this analysis bottleneck by developing chiroptical methods for the ee determination. These chiroptical assays can be conducted in 96-well plates and are conducive to high-throughput experimentation.
One of our multicomponent assays involves an circular dichroism (CD) active octahedral iron complex which incorporates a chiral amine to produce CD out of the absorbance region of common chiral interference compounds. When coupled with a fluorescent indicator displacement assay, both reaction yield and ee can be determined optically and thus in parallel.
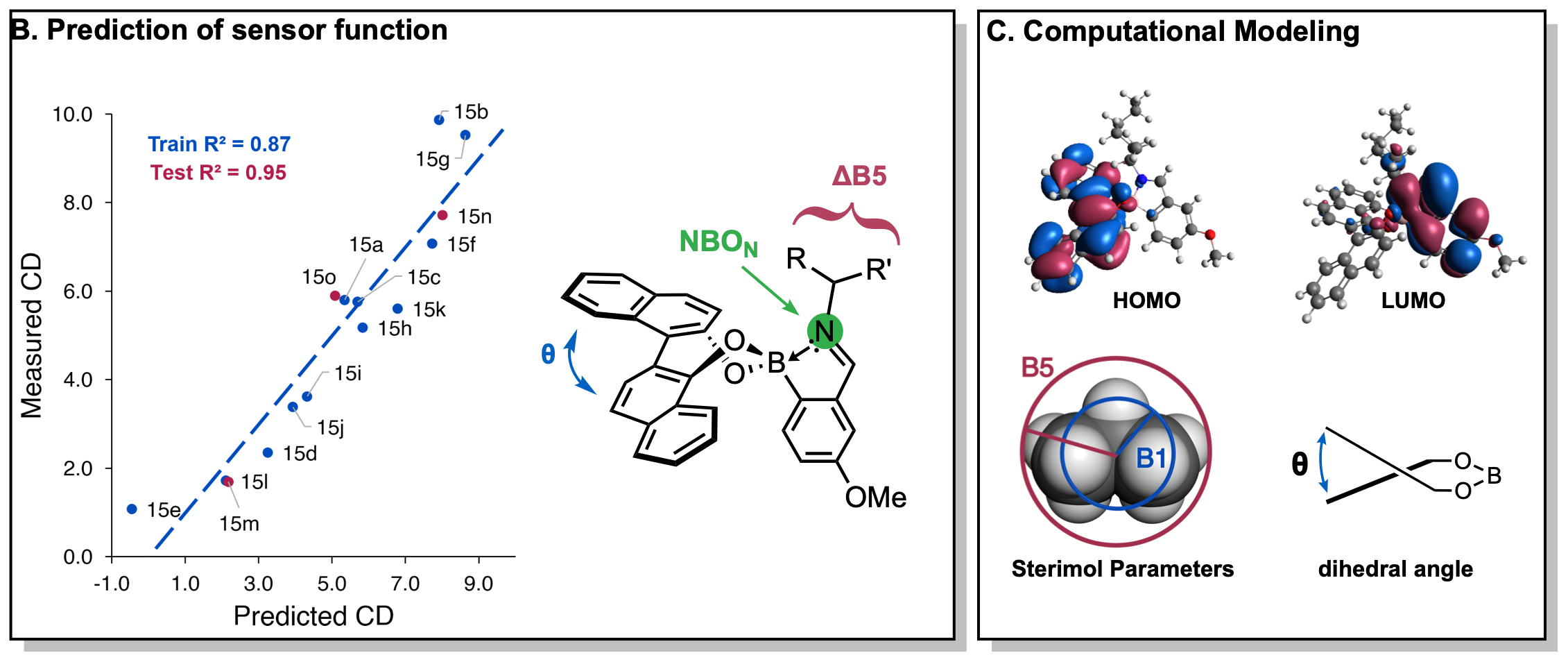
Along with developing chiroptical assays for various functional groups, we’ve parameterized and computationally modeled multicomponent assemblies to understand the physicochemical parameters which govern the CD signal. A predictive model for the CD signal at 100% ee allows operators to circumvent the calibration experiments which relate CD signal to ee further increasing the speed benefits of chiroptical methods over HPLC.
Related Publications:
Tetrahedron 2021, 94, 132315.
More coming soon.
Molecule Sequencing
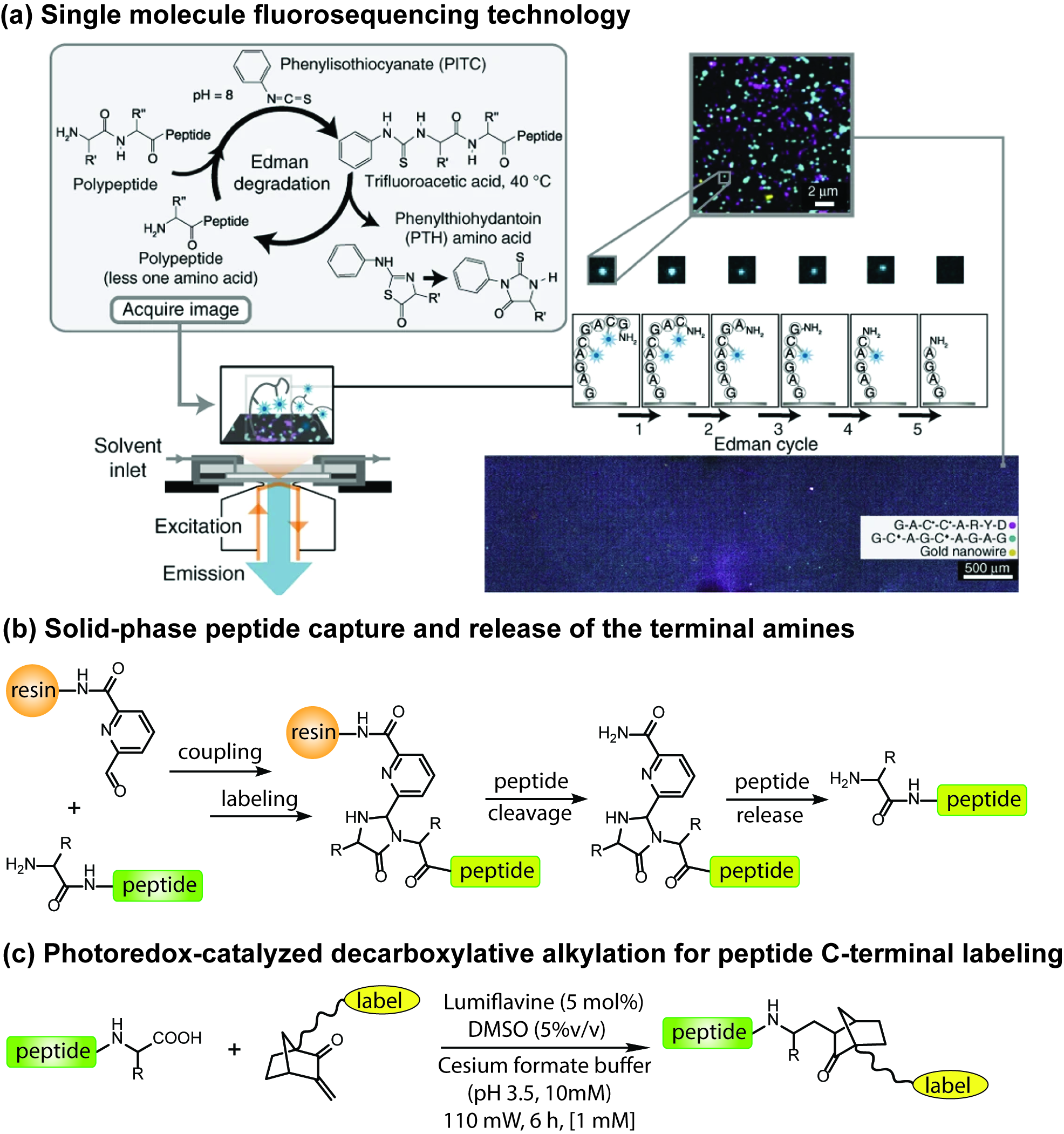
Proteins are biomacromolecules constructed from one or multiple polypeptide chains, which play essential roles in metabolism. The abnormal expression of specific proteins could be taken as one of the symptoms for diseases. However, proteins often exist in very complex environment and a human cell typically contains >10,000 unique proteins which make it extremely challenging to identify and quantify a single molecule in biological samples. Recently, our group, teamed with the Marcotte lab developed a single molecule fluorosequencing technology which offers the capability to identify and quantify every single peptide and protein molecule with low detection limits. The development of new chemical approaches for specific labeling of amino acids and differentiating end groups of peptides are crucial for the application of fluorosequencing technology. To this end, a part of the research in our lab focuses on the following areas: (i) Screening several orthogonal conjugation methods for the efficient labeling of fluorophores; (ii) Developing new chemical methods for the reversible/irreversible capture of N- and C-terminal of peptides; (iii) Exploring mild conditions for Edman degradation, particularly suitable for the fluorosequencing technology; (iv) Discovering chemical approaches to specifically label certain amino acids and peptide sequences. We believe that these chemical tools will not only benefit our fluorosequencing technology but also broadly applicable to other protein and peptide related researches.
Related Publications:
Nat. Biotechnol. 2018, 36, 1076–1082.
ACS Chem. Biol. 2021, 16, 2595–2603.